This is an old revision of the document!
Table of Contents
Working with OMERO
The OMERO server supports multiple front ends to add, download and analyse image data. Currently, there are two o ptions available for OICE users:
- - The web interface accessible through https://romulus.oice.uni-erlangen.de: No configuration is necessary in this case, the only requirement for the user is an existing account in the central user database and an HTML-5 compliant browser.
- - The Java-based OMERO client, available for Windows, Mac OS X and Linux at downloads.openmicroscopy.org/latest/omero5 with its main module, OMERO.insight. This client requires the installation of an up-to-date version of Oracle's Java Runtime Environment. Additionally, the client has to be configured on the first run to find the OICE OMERO server. This will be described in the next chapter.
The web interface is most useful for quick access to image data on third-party computers where no client software is installed. However, certain features like mass downloads of single files or easy drag-and-drop operation are not yet available with the web client (or can not be made available due to inherent limitations of HTML and Javascript), so OMERO.insight would be the preferred option where available.
Configuration of the OMERO client software
- Make sure to first install Java before trying to run the OMERO.insight module.
- - Unzip the software package to an arbitrary directory and run the OMERO.insight executable. You will be presented with a window such as the one below:
.. 
- - Click on the white wrench icon to open the server preferences window:
.. 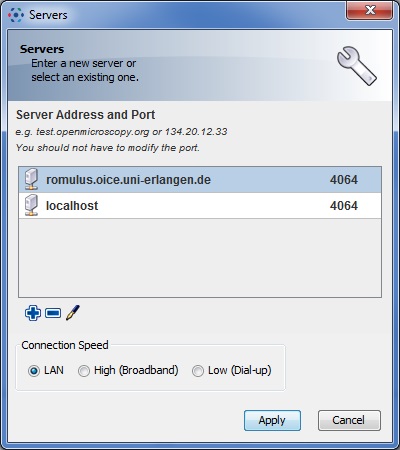
- - Click on the blue plus icon and ad the following server
romulus.oice.uni-erlangen.de. Leave the port (number next to the server) at the default value of4064. - Select the server (by clicking on the name) and click on Apply.
- Next time you open OMERO.insight, the server name left of the white wrench icon should reflect your changes, i.e. state the new server instead of localhost.
- Login with your username and password as usual.
Importing/uploading files to OMERO
There are two ways to upload files to the OMERO server. If you are using one of the OICE's on-site microscopes the preferred way is to use the OMERO DropBox facility. Since the automatic upload has some limitations (files will always appear in the Orphaned Images folder of the last used group), another option is to manually upload your images to the OMERO server. This can be done with the OMERO.insight client, which can both be downloaded separately or called via the OMERO.web interface.
Automatic import via OMERO.DropBox
This is the most comfortable way to upload image files to OMERO. Users at in-house OICE devices (in the future, this will be extended to off-site devices) will be provided with two network shares.
- H:\ which is the home share and has the same name as the user (e.g. mmustermann). Files which are saved here will be accessible on any device but will not be automatically uploaded to OMERO
- X:\ which is the OMERO-DropBox share. If you save any image files on this share, be it on the root level or in any folders or subfolders, those image files will be automatically uploaded to OMERO if the format is recognized (which is the case for over 100 proprietory and open image formats)
Files on the OMERO-DropBox share will not be deleted after upload. Deleting those files is safe and will currently not lead to any action on the OMERO-Server, so your on the server will not be touched once uploaded. The same is true for deleting automatically uploaded files on the server, which will not trigger any action regarding files on your DropBox share. Moving files between folders on the OMERO-Share will not trigger a re-upload, only new files will be uploaded.
<div important>Do not save a file on the DropBox folder before you are finished working with it! If you are planing to add more images to the same file, e.g. by taking more images without starting a new experiment and then pressing Save…, you have to keep your working file outside of the DropBox folder until you have finished your experiment and then copy it over manually. Should you save the same file multiple times inside the DropBox folder, every save-action will trigger a re-upload of the file and you will end up with a lot of identical images. In the worst case, the re-upload will be triggered before the old upload is finished, which can corrupt part of your data.</div>
As soon as the upload is finished (depending on file size, this should not take longer than 5 minutes), users can find the uploaded images in the “Orphaned Images” folder of the last-used group. There is currently no way to indicate the Project or Dataset that automatically uploaded images should end up in, so you have to move those images using the web interface or OMERO.insight.
OMERO.DropBox is a relatively new feature for the OME-Suite, so problems with the automatic upload can still occur. If you find that this feature is not working for you, you can try the manual upload or you can contact the OICE administrator
Manual import via OMERO.insight
Manual import is currently only supported by the Desktop client OMERO.insight. Support for the web interface is planned and will be realized by the OME developers in the near future. Currently, you have to use OMERO.insight, select your target group (in case you are a member of more than one) by using the provided drop-down list and click on the Importer Icon in the top bar to upload your files. A dialog will pop up which allows you to select files, a target Project and a target Dataset.
Downloading files from OMERO
To conduct quantitative and qualitative image analysis that cannot be performed within the OME framework, users have to download the images to the respective computers. This can easily be done with both OMERO.web (the browser interface) and OMERO.insight (the Java client).
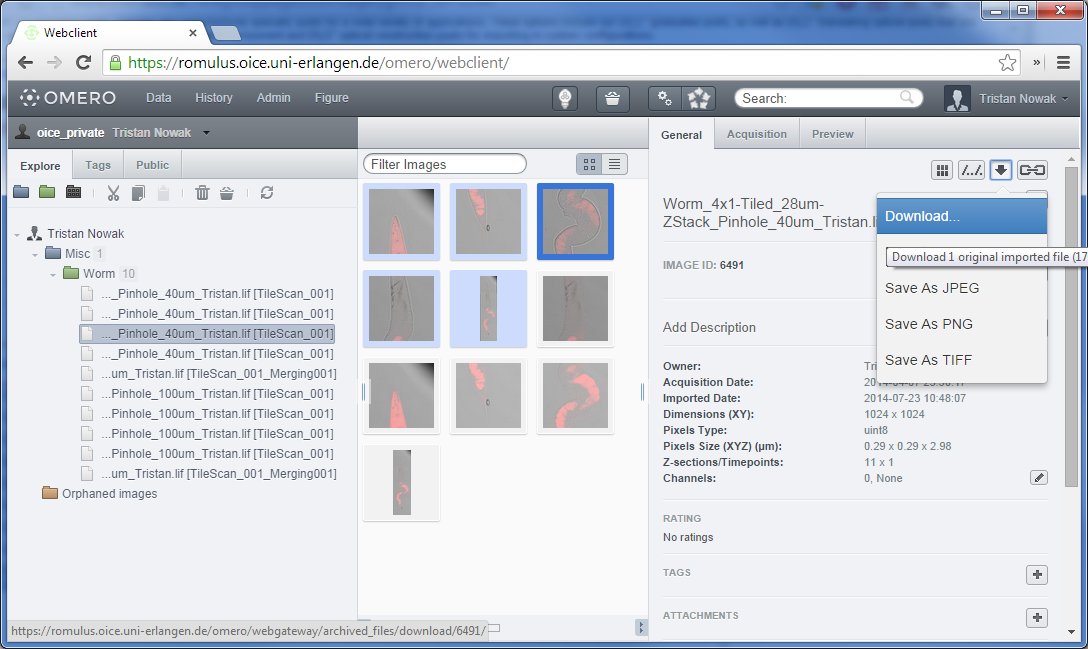
Downloading a single file via OMERO.web
To download a single image via OMERO.web, select the file on the left or center pane and click on the downward arrow button on the right pane as shown above. An option will appear that allows the user do download the original raw data (by just clicking “Download…” as seen in the figure below, or convert the image to various different formats.
Downloading multiple files via OMERO.web
OMERO.web is since version 5.0.3 also capable of easily downloading multiple files. Because of technical limitations of the HTTP protocol, only one file at a time can be downloaded from any web page. To overcome this, OMERO.web packages the files you want to transfer into a ZIP archive that can be extracted with most modern operating systems without any additional software.
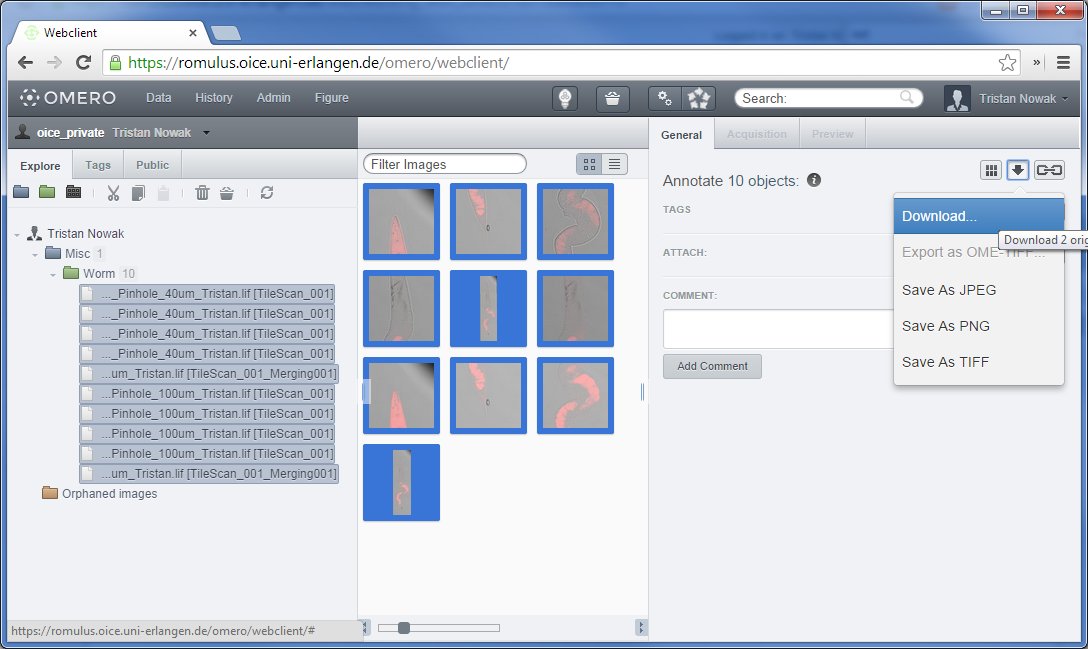
To issue a download for multiple files, select multiple images belonging to multiple raw data files. To do this, select the first image (either on the left or center pane) normally, then hold down SHIFT and select the last one. Alternatively, you can hold down CTRL and click on every single image you would like to transfer. These methods are identical to how multiple file selection is done in Windows explorer.
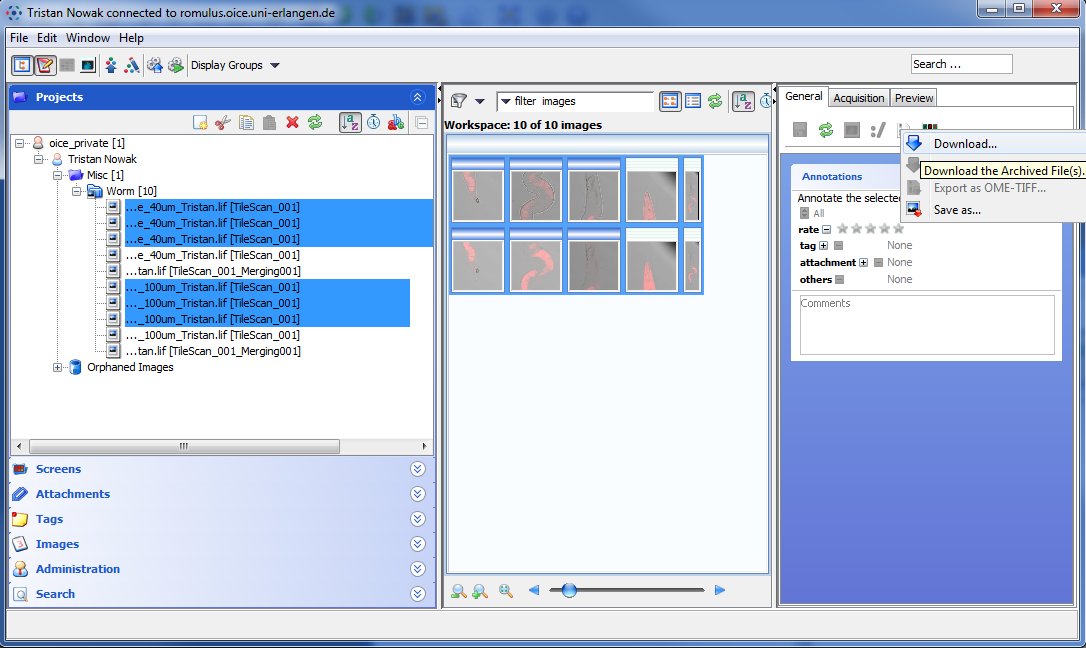
Downlaoding single and multiple files with OMERO.insight
When using OMERO.insight, the limitations of the HTTP protocol do not apply since this client runs locally on your computer. Single and multiple image downloading is handled identically: Select the files you want to download either in the left or center pane and click on the red downward arrow in the right pane as shown in the figure below. Selecting multiple files works identically to OMERO.web, with the additional option of dragging a box around files in the center pane.
If you selected more than one file, you will get a dialog where you can choose the destination folder for all the files. OMERO.insight, just as OMERO.web, supports conversion into a variety of formats before download.
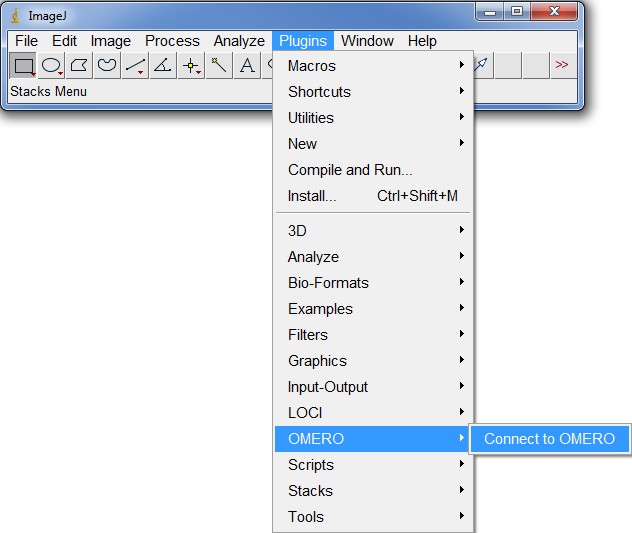
Using ImageJ/Fiji with OMERO
ImageJ is a software for doing a great variety of imaging related data analysis tasks. It can handle multidimensional image data (such as a time series combined with Z-stacks), runs on all platforms thanks to its Java-based implementation and there is a large number of plugins that extent its functionality even further. To simplify the usage of ImageJ for researchers in biological and medical fields, there is an option of using Fiji instead, which is basically ImageJ (or ImageJ2) with a lot of preinstalled plugins, especially for analysis of fluorescence microscopy images. Therefore, for novice users, we recommend installing Fiji over ImageJ.
Both ImageJ and Fiji can be connected to the OMERO server with the same plugin, which allows for a straightforward and easy transfer of acquired microscopy images into these analysis platforms. In the following there will be a short description for new users of how to get both environments running.
For users who want to work with the native MSR format of our STED microscope, it is currently still required to install a beta version of the Bioformats plugin for ImageJ (Fiji will not work with this version, unfortunately). The required daily build can be downloaded here: http://ci.openmicroscopy.org/job/BIOFORMATS-5.0-daily/lastSuccessfulBuild/artifact/artifacts/bioformats_package.jar. It has to be extracted into the plugins\ subdirectory of ImageJ.
Installing ImageJ
Go to http://imagej.nih.gov/ij/download.html or and download the proper version of ImageJ for your system. If you already installed a working OMERO.insight client, you should also already have a working version of Java installed, so you can download the pure version of ImageJ without the bundled Java. The 64bit versions are recommended and should be chosen if you are working on a 64bit system since this will allow you to use more than 3 GB of RAM which is important when working on large data sets.
Installing usually consists of either unzipping the compressed installer packages to a directory of your choice, or running an executable installer and following the instructions on-screen.
Alternatively: Installing Fiji
Goto http://fiji.sc/Downloads and download the proper version of Fiji for your operating system and architecture. As with ImageJ, try to install the 64bit versions if possible. With the exception of Mac OS X, the installer packages are all compressed files and need to be unzipped to a directory of your choice.
Installing the ImageJ/Fiji plugin
Go to downloads.openmicroscopy.org/latest/omero5 and download the ImageJ/Fiji plugin listed under OMERO plugin downloads. As stated above, this plugin works for both ImageJ and Fiji.
To install the plugin, you have to unzip the zip file including all folders to the plugins subdirectory of your ImageJ or Fiji installation directory. For example, if you have installed Fiji in C:\Programs\Fiji, you have to unzip the plugin into C:\Programs\Fiji\plugins. This will lead to a new directory called C:\Programs\Fiji\plugins\OMERO.insight-ij-(version)
After that, you can run ImageJ or Fiji and the plugin should be listed in the top menu under plugins, as shown in the figure below.