Table of Contents
Working with OMERO
The OMERO server supports multiple front ends to add, download and analyse image data. Currently, there are two o ptions available for OICE users:
- - The web interface accessible through https://romulus.oice.uni-erlangen.de: No configuration is necessary. The web interface comes with OMERO.figure for conveniently creating high-quality figures, but cannot be used to import data into the OMERO.server
- - OMERO.insight, a desktop application available for Windows, Mac OS X and Linux at https://github.com/ome/omero-insight/releases/latest. It needs to be configured on the first run to find the OICE OMERO server. This will be described in the next section.
Download, installation and starting of OMERO.insight
- Download OMERO.insight from https://github.com/ome/omero-insight/releases/latest. The .exe file will install it system-wide for all users (admin right required), The .msi file will install it only for the current user (no admin rights required). Mac OS X users should choose the .dmg file.
- If working on a microscope, OMERO.insight is available on the Tools (Y:) drive and can directly started by clicking on the .exe file located within the OMERO.insight folder:

Configuration of OMERO.insight

- - If you are working from within the hospital network, click the lock symbol (2) to encrypt all data transfer
- - Click on the white wrench icon (1) to open the server preferences window. Click on the blue plus icon (1) and add the following server
romulus.oice.uni-erlangen.de, then click on Apply.
.. 
- Next time you open OMERO.insight, the server name left of the white wrench icon should reflect your changes, i.e. state the new server instead of localhost.
- Login with your username and password as usual.
Importing/uploading files to OMERO
- - To start the importer, either select
File>Import…from the menu or click on the importer icon in the tool bar:
- - On the left side, select the files that you want to upload (1) and click the right arrow icon (2):

- - Select a project (1) and a dataset (2) (or choose no project/no dataset, in which case the image will end up under
Orphaned Images) and click onAdd to the queue. You can repeat step 2 and 3 multiple times.
- - After clicking
Import, you'll see the progress of the import. You can close the window once the upload is finished. Back in the OMERO.insight main window, you'll need to click theRefreshbutton to make the new images visible.
Downloading files from OMERO
To conduct quantitative and qualitative image analysis that cannot be performed within the OME framework, users have to download the images to the respective computers. This can easily be done with both OMERO.web (the browser interface) and OMERO.insight (the Java client).
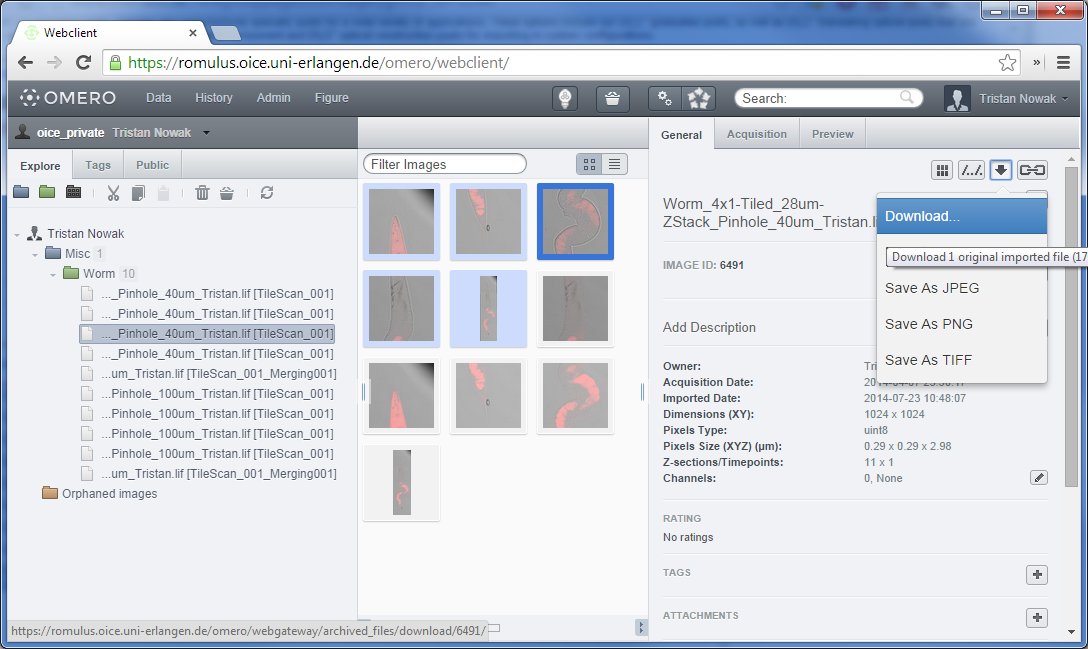
Downloading a single file via OMERO.web
To download a single image via OMERO.web, select the file on the left or center pane and click on the downward arrow button on the right pane as shown above. An option will appear that allows the user do download the original raw data (by just clicking “Download…” as seen in the figure below, or convert the image to various different formats.
Downloading multiple files via OMERO.web
OMERO.web is since version 5.0.3 also capable of easily downloading multiple files. Because of technical limitations of the HTTP protocol, only one file at a time can be downloaded from any web page. To overcome this, OMERO.web packages the files you want to transfer into a ZIP archive that can be extracted with most modern operating systems without any additional software.
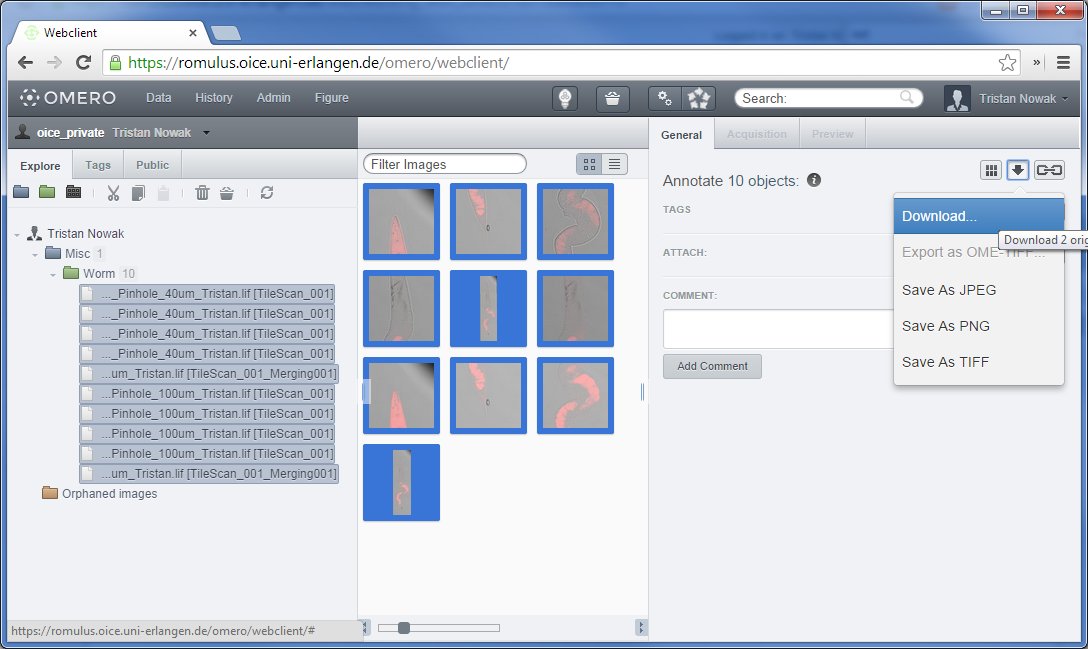
To issue a download for multiple files, select multiple images belonging to multiple raw data files. To do this, select the first image (either on the left or center pane) normally, then hold down SHIFT and select the last one. Alternatively, you can hold down CTRL and click on every single image you would like to transfer. These methods are identical to how multiple file selection is done in Windows explorer.
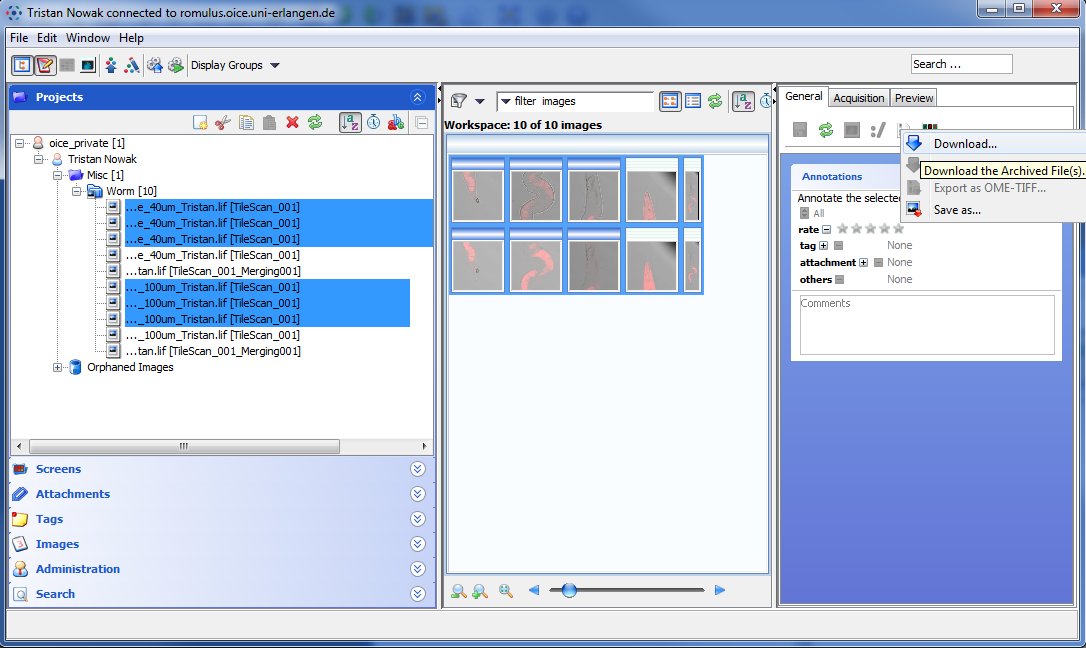
Downlaoding single and multiple files with OMERO.insight
When using OMERO.insight, the limitations of the HTTP protocol do not apply since this client runs locally on your computer. Single and multiple image downloading is handled identically: Select the files you want to download either in the left or center pane and click on the red downward arrow in the right pane as shown in the figure below. Selecting multiple files works identically to OMERO.web, with the additional option of dragging a box around files in the center pane.
If you selected more than one file, you will get a dialog where you can choose the destination folder for all the files. OMERO.insight, just as OMERO.web, supports conversion into a variety of formats before download.
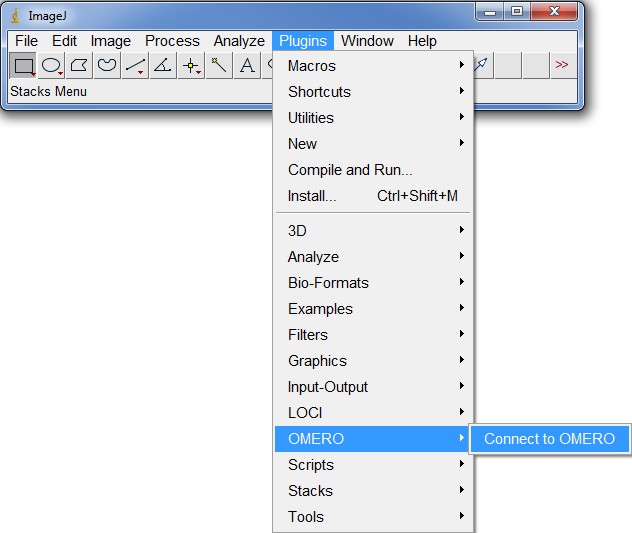
Using ImageJ/Fiji with OMERO
ImageJ is a software for doing a great variety of imaging related data analysis tasks. It can handle multidimensional image data (such as a time series combined with Z-stacks), runs on all platforms thanks to its Java-based implementation and there is a large number of plugins that extent its functionality even further. To simplify the usage of ImageJ for researchers in biological and medical fields, there is an option of using Fiji instead, which is basically ImageJ (or ImageJ2) with a lot of preinstalled plugins, especially for analysis of fluorescence microscopy images. Therefore, for novice users, we recommend installing Fiji over ImageJ.
Both ImageJ and Fiji can be connected to the OMERO server with the same plugin, which allows for a straightforward and easy transfer of acquired microscopy images into these analysis platforms. In the following there will be a short description for new users of how to get both environments running.
For users who want to work with the native MSR format of our STED microscope, it is currently still required to install a beta version of the Bioformats plugin for ImageJ (Fiji will not work with this version, unfortunately). The required daily build can be downloaded here: http://ci.openmicroscopy.org/job/BIOFORMATS-5.0-daily/lastSuccessfulBuild/artifact/artifacts/bioformats_package.jar. It has to be extracted into the plugins\ subdirectory of ImageJ.
Installing ImageJ
Go to http://imagej.nih.gov/ij/download.html or and download the proper version of ImageJ for your system. If you already installed a working OMERO.insight client, you should also already have a working version of Java installed, so you can download the pure version of ImageJ without the bundled Java. The 64bit versions are recommended and should be chosen if you are working on a 64bit system since this will allow you to use more than 3 GB of RAM which is important when working on large data sets.
Installing usually consists of either unzipping the compressed installer packages to a directory of your choice, or running an executable installer and following the instructions on-screen.
Alternatively: Installing Fiji
Goto http://fiji.sc/Downloads and download the proper version of Fiji for your operating system and architecture. As with ImageJ, try to install the 64bit versions if possible. With the exception of Mac OS X, the installer packages are all compressed files and need to be unzipped to a directory of your choice.
Installing the ImageJ/Fiji plugin
Go to https://downloads.openmicroscopy.org/latest/omero5/ and download the ImageJ/Fiji plugin listed under OMERO plugin downloads. As stated above, this plugin works for both ImageJ and Fiji.
To install the plugin, you have to unzip the zip file including all folders to the plugins subdirectory of your ImageJ or Fiji installation directory. For example, if you have installed Fiji in C:\Programs\Fiji, you have to unzip the plugin into C:\Programs\Fiji\plugins. This will lead to a new directory called C:\Programs\Fiji\plugins\OMERO.insight-ij-(version)
After that, you can run ImageJ or Fiji and the plugin should be listed in the top menu under plugins, as shown in the figure below.